Publications
Highlighted Papers
ParSE-seq: a calibrated multiplexed assay to facilitate the clinical classification of putative splice-altering variants Link
Nature Communications (2024).
O’Neill M, Yang T, Laudeman J, Calandranis M, Solus J, Roden D, Glazer A.
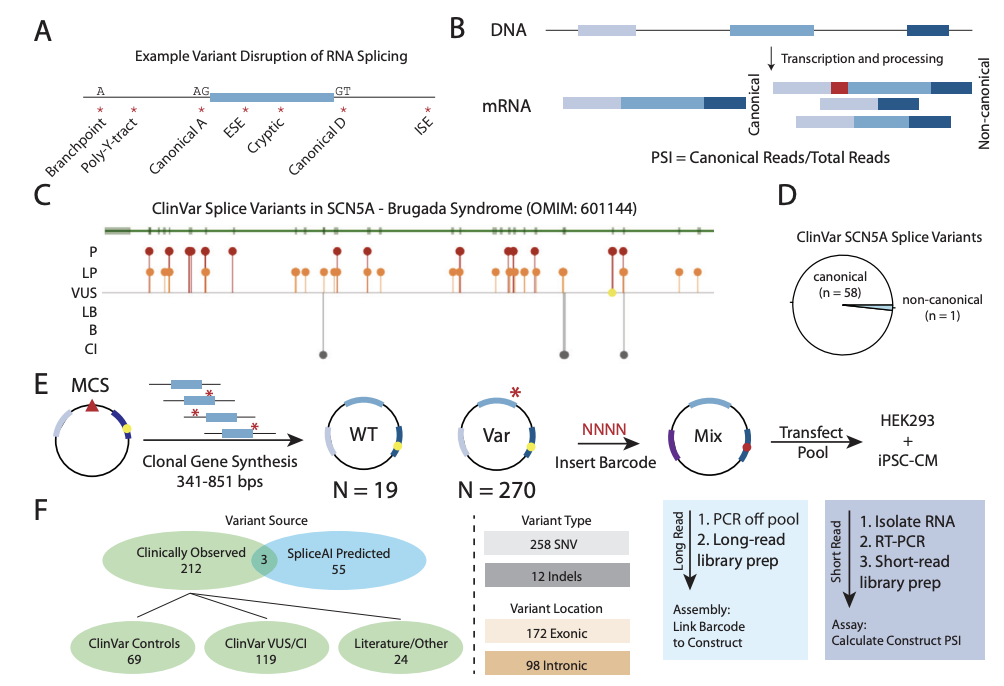
We developed a multiplexed splice assay and studied >200 candidate SCN5A splicing variants in iPS-cardiomyocytes. We validated the assay with lots of benign and pathogenic controls and CRIPSR. Many variants—even coding variants—can have cryptic effects on splicing!
Multi-site validation of a functional assay to adjudicate SCN5A Brugada Syndrome-associated variants. Link
Circulation: Genomic and Precision Medicine (2024).
Ma J, O’Neill M, Richardson E, Thomson K, Ingles J, Muhammad A, Solus J, Davogustto G, Anderson K, Shoemaker M, Stergachis A, Floyd B, Dunn K, Parikh V, Chubb H, Perrin M, Roden D, Vandenberg J*, Ng C*, Glazer A*.
An in vitro SCN5A-BrS automated patch clamp assay was generated for high-throughput functional studies of NaV1.5. The assay was independently studied at two separate research sites – Vanderbilt University Medical Center and Victor Chang Cardiac Research Institute – revealing strong correlations, including peak INa density (R2=0.86). The assay was calibrated according to ClinGen Sequence Variant Interpretation recommendations using high-confidence variant controls (n=49). This validated APC assay provides clinical-grade functional evidence for the reclassification of current VUS and will aid future SCN5A-BrS variant classification.
High-throughput functional mapping of variants in an arrhythmia gene, KCNE1, reveals novel biology Link
Genome Medicine (2024)
Muhammad A, Calandranis M, Li B, Yang T, Blackwell D, Harvey M, Smith J, Chew A, Capra J, Matreyek K, Fowler D, Roden D, Glazer A
We performed a deep mutational scan (comprehensive mutational study) of 2,500 variants in the KCNE1 potassium channel gene. We characterized cell surface trafficking and potassium channel function and discovered hundreds of deleterious variants. The scores were highly concordant with gold standard electrophysiological data, population and patient cohorts, and computational metrics.
Functional Assays Reclassify Suspected Splice-Altering Variants of Uncertain Significance in Mendelian Channelopathies. Pubmed
Circulation: Genomics and Precision Medicine (2022)
O’Neill M, Wada Y, Hall L, Mitchell D, Glazer A*, Roden D.* (*co-corresponding)
We studied 13 variants in cardiac ion channel genes that were near splice sites. We used two assays: CRISPR of induced pluripotent stem cell-derived cardiomyocytes and splicing minigene assays. We found that 9/13 variants had splice-disrupting effects and could be reclassified.
Dominant negative effects of SCN5A missense variants. Pubmed
Genetics in Medicine (2022)
O’Neill M, Muhammad A, Li B, Wada Y, Hall L, Solus J, Short L, Roden D, Glazer A.
Surprisingly, the cardiac ion channel SCN5A was recently shown to dimerize. Here we find widespread dominant negative effects for >30 SCN5A loss of function variants. These variants cause an especially high rate of arrhythmias.
Arrhythmia variant associations and reclassifications in the eMERGE-III sequencing study. Pubmed VUMC Reporter Circulation (2022)
Glazer A, Davogustto G*, Shaffer C, Vanoye C, Desai R, Farber-Eger E, Dikilitas O, Shang N, Pacheco J, Yang T, Muhammad A, Mosley J, Van Driest S, Wells Q, Shaffer L, Kalash O, Wada Y, Bland S, Yoneda Z, Mitchell D, Kroncke B, Kullo I, Jarvik G, Gordon A, Larson E, Manolio T, Mirshahi T, Luo J, Schaid D, Namjou B, Alsaied T, Singh R, Singhal A, Liu C, Weng C, Hripcsak G, Ralston J, McNally E, Chung W, Carrell D, Leppig K, Hakonarson H, Sleiman P, Sohn S, Glessner J, Denny J, Wei W, George A, Shoemaker M, Roden D. *Contributed equally.
What happens when you sequence arrhythmia genes in >20K people? Dozens of pathogenic variants, new diagnoses, and high-throughput in vitro studies.
High-throughput reclassification of SCN5A variants. Pubmed Discover Magazine
Glazer A, Wada Y, Li B, Muhammad A, Kalash O, O’Neill M, Shields T, Hall L, Short L, Blair M, Kroncke B, Capra J, Roden D.
American Journal of Human Genetics (2020). Winner of Cotterman award for paper of the year from a postdoctoral fellow in AJHG.
We used high-throughput automated patch clamping to study the function of 83 variants in the cardiac sodium channel, SCN5A. We found 44 new loss of function variants and reclassified a majority of studied Variants of Uncertain Significance.
Deep Mutational Scan of an SCN5A voltage sensor. Pubmed
Glazer A, Kroncke B, Matreyek K, Yang T, Wada Y, Shields T, Salem J, Fowler D, Roden D.
Circulation Genomic and Precision Medicine (2020). Cover article.
Mutations in the cardiac sodium channel SCN5A can cause arrhythmias, but most are Variants of Uncertain Signficance. We implemented a Deep Mutational Scan of nearly every mutation in a 36-base region of SCN5A. Using high-throughput sequencing and a multiplexed cell death assay, we discovered novel gain and loss of function variants. We are currently scaling this work up to the whole protein.
Full list of publications
Glazer A*, Wilkinson A*, Backer C, Lapan S, Gutzman J, Cheeseman I, Reddien P. The Zn finger protein Iguana impacts Hedgehog signaling by promoting ciliogenesis. Dev Biol. (2010)
Levin T*, Glazer A*, Pachter L, Brem R, Eisen M. Exploring the genetic basis of variation in gene predictions with a synthetic association study. PLoS One (2010)
Miller C*, Glazer A*, Summers B, Blackman B, Norman A, Shapiro M, Cole B, Peichel C, Schluter D, Kingsley D. Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait loci. Genetics (2014)
Glazer A, Cleves P, Erickson P, Lam A, Miller C. Parallel developmental genetic features underlie stickleback gill raker evolution. EvoDevo (2014)
Erickson P, Glazer A, Cleves P, Smith A, Miller C. Two developmentally temporal quantitative trait loci underlie convergent evolution of increased branchial bone length in sticklebacks. Proc Biol Sci. (2014)
Glazer A, Killingbeck E, Mitros T, Rokhsar D, Miller C. Genome Assembly Improvement and Mapping Convergently Evolved Skeletal Traits in Sticklebacks with Genotyping-by-Sequencing. Genes/Genomes/Genetics (G3) (2015)
Ellis N, Glazer A, Donde N, Cleves P, Agoglia R, Miller C. Distinct developmental and genetic mechanisms underlie convergently evolved tooth gain in sticklebacks. Development (2015)
Erickson P, Glazer A, Killingbeck E, Agoglia R, Baek J, Carsanaro S, Lee A, Cleves P, Schluter D, Miller C. Partially repeatable genetic basis of benthic adaptation in threespine sticklebacks. Evolution (2016)
Assad T, Hemnes A, Larkin E, Glazer A, Xu M, Wells Q, Farber-Eger E, Sheng Q, Shyr Y, Harrell F, Newman J, Brittain E. Clinical and Biological Insights Into Combined Post- and Pre-Capillary Pulmonary Hypertension. J Am Coll Cardiology (2016)
Karnes J, Shaffer C, Bastarache L, Gaudieri S, Glazer A, Steiner H, Mosley J, Mallal S, Denny J, Phillips E, Roden D. Comparison of HLA allelic imputation programs. PLoS One (2017)
Karnes J,* Bastarache L,* Shaffer C, Gaudieri S, Xu Y, Glazer A, Mosley J, Zhao S, Raychaudhuri S, Mallal S, Ye Z, Mayer J, Brilliant M, Hebbring S, Roden D, Phillips E, Denny J. Phenome-wide scanning identifies multiple diseases and disease severity phenotypes associated with HLA variants. Science Translational Medicine (2017)
Wells Q, Veatch O, Fessel J, Joon A, Levinson R, Mosley J, Held E, Lindsay C, Shaffer C, Weeke P, Glazer A, Bersell K, Van Driest S, Karnes J, Blair M, Lagrone L, Su Y, Bowton E, Feng Z, Ky B, Lenihan D, Fisch M, Denny J, Roden D. Genome-wide association and pathway analysis of left ventricular function after anthracycline exposure in adults. Pharmacogenetics and Genomics (2017)
Bastarache L, Hughey JJ, Hebbring S, Marlo J, Zhao W, Ho WT, Carroll R, Ramirez AH, Wells QS, Van Driest SL, McGregor TL, Mosley JD, Osterman T, Edwards T, Ruderfer D, Edwards DR, Hamid R, Cogan J, Glazer A, Qi W, Feng Q, Brilliant M, Zhao Z, Cox N, Roden D, Denny J. Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science (2018)
Kroncke B*, Glazer A*, Smith D, Blume J, Roden D. SCN5A (NaV1.5) variant functional perturbation and clinical presentation: variants of a certain significance. Circulation: Genomic and Precision Medicine (2018)
Salem J, Waintraub X. Courtillot C., Schaffer C, Gandjbakhch E, Maupain C, Moslehi J, Badilini F, Haroche J, Gougis P, Fressart V, Glazer A, Hidden-Lucet F, Touraine P, Lebrun-Vignes B, Roden D, Bachelot A, Funck-Brentano C. Hypogonadism as a reversible cause of Torsade de Pointes in men. Circulation (2018)
Johnson C, Potet F, Thompson M, Kroncke B, Glazer A, Voehler M, Knollmann B, George A, Chazin W. A mechanism of calmodulin modulation of the human cardiac sodium channel. Structure (2018).
Grouthier V, Lebrun-Vignes B, Glazer A, Touraine P, Funck-Brentano C, Pariente A, Courtillot C, Bachelot A, Roden D, Moslehi J, Salem J. Increased long QT and Torsade de pointes reporting on tamoxifen compared to aromatase inhibitors. Heart (2018).
Roden D, Glazer A, Kroncke B. Arrhythmia genetics: Not dark and lite, but fifty shades of grey. Heart Rhythm (2018).
Salem J, Shoemaker M, Bastarache L, Shaffer C, Glazer A, Kroncke B, Wells Q, Shi M, Straub P, Jarvik G, Larson E, Edwards D, Edwards T, Davis L, Hakonarson H, Weng C, Fasel D, Knollmann B, Wang T, Denny J, Ellinor P, Roden D, Mosley J. Association of Thyroid Function Genetic Predictors with Atrial Fibrillation. JAMA Cardiology (2019).
Chavali N, Kryshtal D, Parikh S, Wang L, Glazer A, Blackwell D, Kroncke B, Shoemaker M, Knollmann B. Patient-independent human induced pluripotent stem cell model: A new tool for rapid determination of genetic variant pathogenicity in long QT syndrome. Heart Rhythm (2019).
Salem J, Yang T, Moslehi J, Waintraub X, Gandjbakhch E, Bachelot A, Hidden-Lucet F, Hulot J, Knollmann B, Lebrun-Vignes B, Funck-Brentano C, Glazer A, Roden D. Androgenic effects on ventricular repolarization: A translational study from pharmacovigilance databases to iPSC-cardiomyocytes. Circulation (2019).
eMERGE Consortium (Zouck H, ... Glazer A, … et al). Harmonizing Clinical Sequencing and Interpretation for the eMERGE III Network. American Journal of Human Genetics (2019).
Glazer A, Kroncke B, Matreyek K, Yang T, Wada Y, Shields T, Salem J, Fowler D, Roden D. Deep Mutational Scan of an SCN5A voltage sensor. Circulation Genomic and Precision Medicine (2020). Cover article.
Glazer A, Wada Y, Li B, Muhammad A, Kalash O, O’Neill M, Shields T, Hall L, Short L, Blair M, Kroncke B, Capra J, Roden D. High-throughput reclassification of SCN5A variants. American Journal of Human Genetics (2020). Winner of Cotterman award for paper of the year from a postdoctoral fellow in AJHG.
Kozek K*, Glazer A*, Ng C, Blackwell D, Egly C, Vanags L, Blair M, Mitchell D, Matreyek K, Fowler D, Knollmann B, Vandenberg J, Roden D, Kroncke B. High-throughput discovery of trafficking-deficient variants in the cardiac potassium channel KV11.1. Heart Rhythm (2020).
Kroncke B, Smith D, Zuo Y, Glazer A, Roden D, Blume J. A Bayesian method to estimate variant-induced disease penetrance. PLOS Genetics (2020).
Streeten E, See V, Jeng L, Maloney K, Lynch M, Glazer A, Yang T, Roden D, Pollin T, Daue M, Ryan K, Perry J, O’Connell J, Beitelshees A, Palmer K, Mitchell B, Shuldiner A. KCNQ1 and Long QT Syndrome in 1/45 Amish: The road from identification to implementation of culturally appropriate precision medicine. Circulation: Genomic and Precision Medicine (2020).
Yoneda Z, Anderson K, Quintana J, O’Neill M, Sims R, Glazer A, Shaffer C, Crawford D, Stricker T, Ye F, Wells Q, Stevenson L, Michaud G, Darbar D, Lubitz S, Ellinor P, Roden D, Shoemaker B. Early-onset Atrial Fibrillation and the Prevalence of Rare Variants in Cardiomyopathy and Arrhythmia Genes. JAMA: Cardiology (2021).
Kozek K, Wada Y, Sala L, Denjoy I, Egly C, O’Neill M, Aiba T, Shimizu W, Makita N, Ishikawa T, Crotti L, Spazzolini C, Kotta M, Dagradi F, Castelletti S, Pedrazzini M, Gnecchi M, Leenhardt A, Salem J, Ohno S, Zuo Y, Glazer A, Mosley J, Roden D, Knollman B, Blume J, Extramiana F, Schwartz P, Horie M, Kroncke B. Estimating the Post-Test Probability of Long QT Syndrome Diagnosis for Rare KCNH2 Variants. Circulation: Genomic and Precision Medicine (2021).
Papagiannis J, Yang T, Glazer A, Tisma-Dupanovic S, Avramidis D, Kannankeril P, Viskin S, Walsh E, Roden D. Incessant Atrial and Ventricular Tachycardias associated with an SCN5A mutation. Heart Rhythm Case Reports (2021).
Glazer A*, Davogustto G*, Shaffer C, Vanoye C, Desai R, Farber-Eger E, Dikilitas O, Shang N, Pacheco J, Yang T, Muhammad A, Mosley J, Van Driest S, Wells Q, Shaffer L, Kalash O, Wada Y, Bland S, Yoneda Z, Mitchell D, Kroncke B, Kullo I, Jarvik G, Gordon A, Larson E, Manolio T, Mirshahi T, Luo J, Schaid D, Namjou B, Alsaied T, Singh R, Singhal A, Liu C, Weng C, Hripcsak G, Ralston J, McNally E, Chung W, Carrell D, Leppig K, Hakonarson H, Sleiman P, Sohn S, Glessner J, the eMERGE Network; Denny J, Wei W, George A, Shoemaker M, Dan M. Roden D. Arrhythmia variant associations and reclassifications in the eMERGE-III sequencing study. Circulation (2022).
Wada Y, Yang T, Shaffer C, Daniel L, Glazer A, Davogustto G, Lowery B, Farber-Eger E, Wells Q, Roden D. Common ancestry-specific ion channel variants predispose to drug-induced arrhythmias. Circulation (2022).
O’Neill M, Muhammad A, Li B, Wada Y, Hall L, Solus J, Short L, Roden D*, Glazer A*. Dominant negative effects of SCN5A missense variants. Genetics in Medicine (2022).
Gulsevin A, Glazer A, Shields T, Kroncke B, Roden D, Meiler J. Veratridine Can Bind to a Site at the Mouth of the Channel Pore at Human Cardiac Sodium Channel NaV1.5. International Journal of Molecular Sciences (2022).
Yoneda Z, Anderson K, Ye F, Quintana J, O’Neill M, Sims R, Sun L, Glazer A, Davogustto G, El-Harasis M, Laws J, Saldivar B, Crawford D, Stricker T, Wells Q, Darbar D, Michaud G, Stevenson L, Lubitz S, Ellinor P, Roden D, Shoemaker B. Mortality among Patients with Early-onset Atrial Fibrillation and Rare Variants in Cardiomyopathy and Arrhythmia Genes. JAMA Cardiology (2022).
O’Neill M, Wada Y, Hall L, Mitchell D, Glazer A*, Roden D* (Co-corresponding authors). Functional Assays Reclassify Suspected Splice-Altering Variants of Uncertain Significance in Mendelian Channelopathies. Circulation: Genomic and Precision Medicine (2022).
Glazer A. Genetics of Congenital Arrhythmia Syndromes: The Challenge of Variant Interpretation. Current Opinion in Genetics and Development (2022).
Bersell K, Yang T, Mosley J, Glazer A, Hale A, Kryshtal D, Kim K, Steimle J, Brown J, Salem J, Campbell C, Hong C, Wells Q, Johnson A, Short L, Blair M, Behr E, Petropoulou E, Jamshidi Y, Benson M, Keyes M, Ngo D, Vasan R, Yang Q, Gerszten R, Shaffer C, Parikh S, Sheng Q, Kannankeril P, Moskowitz I, York J, Wang T, Knollmann B, and Roden D. Transcriptional dysregulation underlies both a monogenic arrhythmia syndrome and common modifier of cardiac repolarization. Circulation (2023).
Floyd B, Weile J, Kannakeril P, Glazer A, Reuter C, MacRae C, Ashley E, Roden D, Roth F, and Parikh V. Proactive variant effect mapping aids diagnosis in pediatric cardiac arrest. Circulation: Genomics and Precision Medicine (2023).
O’Neill M, Chen S, Rumping L, Johnson R, van Slegtenhorst M, Glazer A, Yang T, Solus J, Laudeman J, Mitchell D, Vanags L, Kroncke B, Anderson K, Gao S, Verdonschot J, Brunner H, Hellebrekers D, Taylor M, Roden D, Wessels M, Deprez R, Fatkin D, Mestroni L, Shoemaker B. Multicenter Clinical and Functional Evidence Reclassifies a Recurrent Non-canonical FLNC Splice-altering Variant. Heart Rhythm (2023).
Wada Y, Wang L, Hall L, Yang T, Short L, Solus J, Glazer A, Roden D. The electrophysiologic effects of KCNQ1 extend beyond expression of IKs: evidence from genetic and pharmacologic block. Cardiovascular Research (2024).
Claussnitzer M, Parikh V, Wagner A, Arbesfeld J, Bult C, Firth H, Muffley L, Nguyen Ba A, Riehle K, Roth F, Tabet D, Bolognesi B*, Glazer A*, Rubin A*. Minimum information and guidelines for reporting a Multiplexed Assay of Variant Effect. Genome Biology (2024).
Muhammad A, Calandranis M, Li B, Yang T, Blackwell D, Harvey M, Smith J, Daniel Z, Chew A, Capra J, Matreyek K, Fowler D, Roden D, Glazer A. High-throughput functional mapping of variants in an arrhythmia gene, KCNE1, reveals novel biology. Genome Medicine (2024).
Ma J, O’Neill M, Richardson E, Thomson K, Ingles J, Muhammad A, Solus J, Davogustto G, Anderson K, Shoemaker M, Stergachis A, Floyd B, Dunn K, Parikh V, Chubb H, Perrin M, Roden D, Vandenberg J*, Ng C*, Glazer A*. Multi-site validation of a functional assay to adjudicate SCN5A Brugada Syndrome-associated variants. Circulation Gen. Prec. Med. (2024).
O’Neill M, Ng C, Aizawa T, Sala L, Bains S, Winbo A, Ullah R, Shen Q, Tan C, Kozek K, Vanags L, Mitchell D, Shen A, Wada Y, Kashiwa A, Crotti L, Dagradi F, Musu G, Spazzolini C, Neves R, Bos J, Giudicessi J, Bledsoe X, Gamazon E, Lancaster M, Glazer A, Knollmann B, Roden D, Weile J, Roth F, Salem J, Earle N, Stiles R, Agee T, Johnson C, Horie M, Skinner J, Ackerman M, Schwartz P, Ohno S, Vandenberg J, Kroncke B. Assays of Variant Effect and Automated Patch Clamping Improve KCNH2-LQTS Variant Classification and Cardiac Event Risk Stratification. Circulation (2024).
O’Neill M, Yang T, Laudeman J, Calandranis M, Solus J, Roden D, Glazer A. ParSE-seq: A Calibrated Multiplexed Assay to Facilitate the Clinical Classification of Putative Splice-altering Variants. Nature Communications (2024).
Patel N, Shen A, Wada Y, Blair M, Mitchell D, Vanags L, Woo S, Ku M, Dauda K, Morris W, Yang M, Knollmann B, Salem J, Glazer A,* Kroncke B*. (* co-corresponding). A High-Performance Extracellular Field Potential Analyzer for iPSC-Derived Cardiomyocytes. Scientific Reports (2025).